CTFS Tutorials
Source file: http://ctfs.si.edu/ctfsdev/CTFSRPackageNew/files/tutorials/abundFitModelPopulation Changes
FLUCTUATION IN ABUNDANCE OF TREE SPECIES
Date: August 13, 2012.
The question addressed here is how much abundances of all the tree species in a plot fluctuate from census to census. At one extreme, abundances are stabilized, and populations do not change. In contrast, there may be environmental drivers favoring some species and not others, so that some species show marked (ie, more than neutral) increases and others decreases. In between the two extremes is a community in which no external drivers affect abundances but populations fluctuate due to demographic stochasticity: a neutral forest.
The test of these models hinges on a histogram of the rate of population change of every species in the
forest. Define λ = 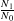 , the ratio of a future population N1 to the current N0, and r = little.r = logλ.
In a highly stabilized community, λ ≈ 1 for every species, and r = 0, whereas in a forest with
environmental fluctionas, λ may be distributed widely but ought to be centered close to 1.
The neutral community would fall in between, with λ varying a small amount due to chance
alone.
, the ratio of a future population N1 to the current N0, and r = little.r = logλ.
In a highly stabilized community, λ ≈ 1 for every species, and r = 0, whereas in a forest with
environmental fluctionas, λ may be distributed widely but ought to be centered close to 1.
The neutral community would fall in between, with λ varying a small amount due to chance
alone.
The distribution of λ and r under neutrality can be predicted mathematically, assuming deaths are binomially distributed across individuals and a Poisson distribution of births whose mean matches the death rate. Because of this statistical model of neutral fluctuations, any environmentally caused variation can be estimated precisely. In a conceptual sense, it means subtracting the predicted neutral variance of λ from the observed variance of λ; what’s left is the environmental variances. The neutral prediction depends on the mortality rate, which is also fitted by the model. The following demonstrates an R program for running this model with forest plot abundances.
The model allows various functional forms for the distribution of population growth. The example below is based on a power probability distribution, where p(x) ∼ x + 1k (k < −1). The exponent k describes how broad the distribution, with more extreme k (ie, more negative) meaning a narrower distribution. Indeed, frac1k is close to the standard deviation of the distribution of r. The power distribution, though, allows k on the negative size of x to differ from k for x positive. It is thus an asymmetric power distribution.
1. Formatting data
The calculations start with the full plot dataframes described at http://ctfs.arnarb.harvard.edu/Public/CTFSRPackage/help/RoutputFull.pdf. The BCI data can be downloaded from http://ctfs.arnarb.harvard.edu/bci/RAnalyticalTables after filling out the request form http://ctfs.arnarb.harvard.edu/webatlas/datasets/bci/. Assume for the following example that dataframes for three censues are loaded, bci.full1, bci.full2, and bci.full3. A species table, such as bci.spptable, is also needed. The species table has a column IDlevel that indicates which taxa are neither valid species nor valid morphospecies – cases where a species code includes a mixture of unknown species. The typical example of these mixtures are unidentified trees, with codes such as UNID or uniden. They must be excluded from the analysis, since changes in abundance of those species codes do not represent population fluctuations of a species.
1 1 1 -05599 swars1 4007 800.2 152.2 1 1 90 1.3 1.3 1981-06-12 alive <NA> 1 7833.000 A 0.0426371077
2 2 1 -22851 hybapr 0718 151.5 378.8 2 1 35 1.3 1.3 1982-07-01 alive L 1 8217.000 A 0.0028894861
3 3 1 -24362 aegipa 0417 95.2 357.5 3 1 10 1.3 1.3 1982-07-02 alive <NA> 1 8218.000 A 0.0001466688
4 4 NA -26589 <NA> beilpe 0007 11.7 151.1 NA NA NA <NA> NA <NA> prior <NA> NA 8237.215 P 0.0000000000
5 5 NA -26590 <NA> faraoc 0004 7.7 96.2 NA NA NA <NA> NA <NA> prior <NA> NA 8233.467 P 0.0000000000
6 6 NA -26703 <NA> hybapr 0210 50.1 215.4 NA NA NA <NA> NA <NA> prior <NA> NA 8234.400 P 0.0000000000
1 1 1 -05599 swars1 4007 800.2 152.2 1 2 94 1.3 1.3 1985-06-28 alive Z 1 9310 A 0.0478479558
2 2 1 -22851 hybapr 0718 151.5 378.8 2 2 NA <NA> NA 1985-03-06 dead D,MC 1 9196 D 0.0000000000
3 3 1 -24362 aegipa 0417 95.2 357.5 3 2 15 1.3 1.3 1985-02-06 alive <NA> 1 9168 A 0.0003618661
4 4 1 -26589 beilpe 0007 11.7 151.1 4 2 10 1.3 1.3 1985-01-11 alive Z 1 9142 A 0.0001132402
5 5 1 -26590 faraoc 0004 7.7 96.2 5 2 10 1.3 1.3 1985-01-08 alive Z 1 9139 A 0.0001313070
6 6 1 -26703 hybapr 0210 50.1 215.4 6 2 10 1.3 1.3 1985-01-23 alive Z 1 9154 A 0.0001496468
1 1 1 -05599 swars1 4007 800.2 152.2 1 3 NA <NA> NA 1990-10-20 dead D,N 1 11250 D 0
2 2 NA -22851 <NA> hybapr 0718 151.5 378.8 NA NA NA <NA> NA <NA> dead <NA> NA 11135 D 0
3 3 1 -24362 aegipa 0417 95.2 357.5 2 3 NA <NA> NA 1990-04-06 dead D,N 1 11053 D 0
4 4 1 -26589 beilpe 0007 11.7 151.1 3 3 NA <NA> NA 1990-02-23 dead D,N 1 11011 D 0
5 5 1 -26590 faraoc 0004 7.7 96.2 4 3 NA <NA> NA 1990-02-16 dead D,N 1 11004 D 0
6 6 1 -26703 hybapr 0210 50.1 215.4 5 3 NA <NA> NA 1990-03-07 dead D,N 1 11023 D 0
acac1 acac1 Acacia sp.1 Acacia sp.1 Fabaceae-mimosoideae 5 genus <NA>
acacme acacme Acacia melanoceras Acacia melanoceras Fabaceae-mimosoideae 4 Beurl. species <NA>
acacri acacri Acacia riparia Acacia riparia Fabaceae-mimosoideae 1192 Kunth species <NA>
acaldi acaldi Acalypha diversifolia Acalypha diversifolia Euphorbiaceae 6 Jacq. species <NA>
acalma acalma Acalypha macrostachya Acalypha macrostachya Euphorbiaceae 7 Jacq. species <NA>
ade1tr ade1tr Adelia triloba Adelia triloba Euphorbiaceae 12 (Müll.Arg.) Hemsl. species <NA>
2. Running the model of environmental variance
A function in the CTFSRPackage (topic population change) called model.littleR.Gibbs runs the model to estimate leftover environmental variance over two census intervals. To run with your own data, simply replace census1 and census2 with your own R Analytical Tables for a CTFS plot. The fitting procedure requires a Bayesian parameter-search, which must run at least 1000 steps. I recommend here running the model once for 1200 steps, as in the example below in order to explore results. In order to get really solid parameter estimates, a run of 10,000 steps should be set. Ddepending on your computer, 1000 steps should take about 3 minutes and 10,000 steps 30 minutes. Note that mindbh needs to be set. The example below assumes dbh is in mm, and is set to 10.
+ bad.modelparam=bad.asympower.param,steps=1200,burn=200,showstep=25)
3. Model results
The result, saved object as mod, is a Bayesian model object including the estimated parameters and their confidence, plus the complete parameter search. The element of the list mean has the fitted parameters. The first two describe the estimated distribution of mortality rates:
- hyperMu = mean of the log(mortality) rate across species;
- hyperSD = standard deviation of the log(mortality) rate across species.
The next three parameters describe the distribution of population growth rates, as defined by r = logλ:
- hyperR = mean of r across species, generally very close to 0;
- hyperSDlow = inverse of the exponent of the power distribution for x < 0; its negative is similar in magnitude to the median population population change of those species declining in abundance;
- hyperSDup = IBID for the positive half of x; its negative is similar in magnitude to the median population population change of those species increasing in abundance.
Upper and lower confidence limits for the five parameters are returned in the components upper and lower.
-3.128642757 0.754418836 0.002269805 -0.037181027 -0.028431362
-3.06894740 0.80445746 0.01407869 -0.02976379 -0.02522590
-3.207321654 0.680282454 -0.002896329 -0.086797191 -0.032366787
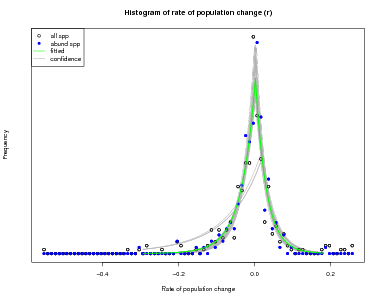
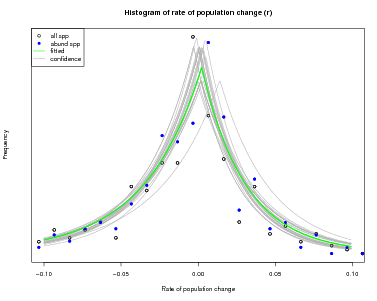
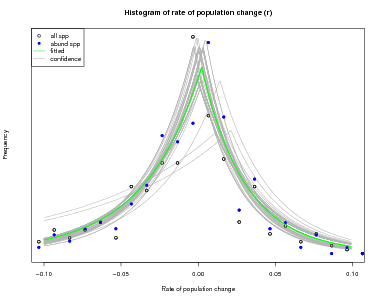
There is another function in the same topic of the CTFSRPackage called graph.abundmodel that creates a graphical view of the fitted model. Pass it the name of the model object and nothing more and it will show a histogram of observed rates of population change r (open circles for all species, blue circles for abundant species), along with the fit to the histogram (green line).
The green curve is the estimated variation in in abundance due to the environment: what is left over after stochastic demography is accounted for. The points, which give the observed histogram, include stochastic plus environmental, thus should be broader than the green curve.
The x-limits of the graph can be adjusted using the argument xrange. In addition, confidence limits can be added by setting conf=50, or any other positive number (the number of lines to draw). The confidence lines are based on all the steps in the Bayesian sampling and reflect confidence in the fitted model.
N1 N2 S time date1 date2 mortrate little.r fitmort lowermean uppermean fitr lowermeanR uppermeanR
paligu 376 661 247 3.053886 8105.625 9228.600 0.137595454 0.18473667 0.13500518 0.109353135 0.15631900 0.18048402 0.14736788 0.20665624
clidde 8 14 7 2.796704 8068.595 9256.214 0.047745989 0.20009834 0.04783252 0.020310979 0.12975013 0.09787158 0.01551216 0.19277640
ochrpy 4 8 3 2.691700 8181.110 9181.840 0.106877479 0.25751282 0.06501124 0.026810071 0.16409626 0.09578779 0.01835821 0.26812851
ingath 61 78 58 2.649370 8196.134 9185.458 0.019035038 0.09278998 0.02411662 0.010499432 0.04939158 0.07838768 0.03188894 0.12057637
micoar 528 677 389 3.195909 8054.420 9251.310 0.095596265 0.07777913 0.09573952 0.082168073 0.10559762 0.07026545 0.05952112 0.08733782
cuparu 56 73 49 3.273421 8080.136 9244.732 0.040792615 0.08098799 0.04987732 0.022473972 0.08699604 0.06975892 0.02503962 0.10718172
cha2sc 194 239 190 2.895034 8160.319 9220.697 0.007196491 0.07205628 0.01108164 0.004770191 0.02242886 0.06940662 0.05064604 0.09660343
urerba 2 5 0 3.812822 7954.932 9276.985 Inf 0.24031825 0.13493791 0.022634915 0.35041866 0.06671173 -0.02216408 0.18462937
myrcga 40 50 38 3.231830 8054.830 9266.683 0.015871284 0.06904557 0.02366960 0.009351584 0.04020493 0.06306927 0.02731841 0.10487277
eugega 963 1162 917 3.345195 8044.492 9249.944 0.014631716 0.05615354 0.01599438 0.012605254 0.01824705 0.05595504 0.04743933 0.06617516
$Biggest_losses
N1 N2 S time date1 date2 mortrate little.r fitmort lowermean uppermean fitr lowermeanR uppermeanR
bactc1 241 84 84 3.500939 8002.949 9273.566 0.30105644 -0.30105644 0.29367749 0.26478207 0.3297441 -0.29300499 -0.3248475 -0.22733664
bactba 111 55 55 3.388399 8024.983 9265.000 0.20723565 -0.20723565 0.19885841 0.15353804 0.2639641 -0.19553875 -0.2600445 -0.12258659
pipecu 120 65 62 3.012689 8119.954 9219.893 0.21919198 -0.20350736 0.20422073 0.15152511 0.2847200 -0.18373390 -0.2359292 -0.09849162
bactc2 38 17 17 3.290464 8056.410 9252.538 0.24445571 -0.24445571 0.20608992 0.12881790 0.3019426 -0.17023845 -0.2763264 -0.04498295
hampap 76 49 41 3.217818 8010.668 9258.138 0.19179499 -0.13640085 0.17426848 0.12649105 0.2490157 -0.10871699 -0.1640417 -0.03893491
cecrob 61 38 27 3.123925 8036.937 9260.472 0.26090156 -0.15150417 0.24426914 0.17270408 0.3425667 -0.10431432 -0.1706922 -0.03689557
sennda 201 135 114 3.363347 8028.548 9262.585 0.16861370 -0.11834345 0.16496953 0.14534654 0.2020067 -0.10397926 -0.1475743 -0.05487117
bactma 469 355 341 2.993622 8130.976 9225.702 0.10646643 -0.09302609 0.10539174 0.09120041 0.1183469 -0.08910109 -0.1066423 -0.06859027
conoci 389 279 232 3.555180 7989.365 9270.768 0.14537716 -0.09348825 0.14290808 0.12359480 0.1677055 -0.08625363 -0.1112317 -0.05920075
turpoc 153 113 112 3.229882 8027.949 9258.576 0.09657909 -0.09382700 0.09159398 0.06736020 0.1227418 -0.08600706 -0.1123598 -0.05152248
N1 N2 S time date1 date2 mortrate little.r fitmort lowermean uppermean fitr lowermeanR uppermeanR
paligu 376 661 247 3.053886 8105.625 9228.600 0.137595454 0.18473667 0.13500518 0.109353135 0.15631900 0.18048402 0.14736788 0.20665624
clidde 8 14 7 2.796704 8068.595 9256.214 0.047745989 0.20009834 0.04783252 0.020310979 0.12975013 0.09787158 0.01551216 0.19277640
ochrpy 4 8 3 2.691700 8181.110 9181.840 0.106877479 0.25751282 0.06501124 0.026810071 0.16409626 0.09578779 0.01835821 0.26812851
ingath 61 78 58 2.649370 8196.134 9185.458 0.019035038 0.09278998 0.02411662 0.010499432 0.04939158 0.07838768 0.03188894 0.12057637
micoar 528 677 389 3.195909 8054.420 9251.310 0.095596265 0.07777913 0.09573952 0.082168073 0.10559762 0.07026545 0.05952112 0.08733782
cuparu 56 73 49 3.273421 8080.136 9244.732 0.040792615 0.08098799 0.04987732 0.022473972 0.08699604 0.06975892 0.02503962 0.10718172
cha2sc 194 239 190 2.895034 8160.319 9220.697 0.007196491 0.07205628 0.01108164 0.004770191 0.02242886 0.06940662 0.05064604 0.09660343
urerba 2 5 0 3.812822 7954.932 9276.985 Inf 0.24031825 0.13493791 0.022634915 0.35041866 0.06671173 -0.02216408 0.18462937
myrcga 40 50 38 3.231830 8054.830 9266.683 0.015871284 0.06904557 0.02366960 0.009351584 0.04020493 0.06306927 0.02731841 0.10487277
eugega 963 1162 917 3.345195 8044.492 9249.944 0.014631716 0.05615354 0.01599438 0.012605254 0.01824705 0.05595504 0.04743933 0.06617516
$Biggest_losses
N1 N2 S time date1 date2 mortrate little.r fitmort lowermean uppermean fitr lowermeanR uppermeanR
bactc1 241 84 84 3.500939 8002.949 9273.566 0.30105644 -0.30105644 0.29367749 0.26478207 0.3297441 -0.29300499 -0.3248475 -0.22733664
bactba 111 55 55 3.388399 8024.983 9265.000 0.20723565 -0.20723565 0.19885841 0.15353804 0.2639641 -0.19553875 -0.2600445 -0.12258659
pipecu 120 65 62 3.012689 8119.954 9219.893 0.21919198 -0.20350736 0.20422073 0.15152511 0.2847200 -0.18373390 -0.2359292 -0.09849162
bactc2 38 17 17 3.290464 8056.410 9252.538 0.24445571 -0.24445571 0.20608992 0.12881790 0.3019426 -0.17023845 -0.2763264 -0.04498295
hampap 76 49 41 3.217818 8010.668 9258.138 0.19179499 -0.13640085 0.17426848 0.12649105 0.2490157 -0.10871699 -0.1640417 -0.03893491
cecrob 61 38 27 3.123925 8036.937 9260.472 0.26090156 -0.15150417 0.24426914 0.17270408 0.3425667 -0.10431432 -0.1706922 -0.03689557
sennda 201 135 114 3.363347 8028.548 9262.585 0.16861370 -0.11834345 0.16496953 0.14534654 0.2020067 -0.10397926 -0.1475743 -0.05487117
bactma 469 355 341 2.993622 8130.976 9225.702 0.10646643 -0.09302609 0.10539174 0.09120041 0.1183469 -0.08910109 -0.1066423 -0.06859027
conoci 389 279 232 3.555180 7989.365 9270.768 0.14537716 -0.09348825 0.14290808 0.12359480 0.1677055 -0.08625363 -0.1112317 -0.05920075
turpoc 153 113 112 3.229882 8027.949 9258.576 0.09657909 -0.09382700 0.09159398 0.06736020 0.1227418 -0.08600706 -0.1123598 -0.05152248
N1 N2 S time date1 date2 mortrate little.r fitmort lowermean uppermean fitr lowermeanR uppermeanR
paligu 376 661 247 3.053886 8105.625 9228.600 0.137595454 0.18473667 0.13500518 0.109353135 0.15631900 0.18048402 0.14736788 0.20665624
clidde 8 14 7 2.796704 8068.595 9256.214 0.047745989 0.20009834 0.04783252 0.020310979 0.12975013 0.09787158 0.01551216 0.19277640
ochrpy 4 8 3 2.691700 8181.110 9181.840 0.106877479 0.25751282 0.06501124 0.026810071 0.16409626 0.09578779 0.01835821 0.26812851
ingath 61 78 58 2.649370 8196.134 9185.458 0.019035038 0.09278998 0.02411662 0.010499432 0.04939158 0.07838768 0.03188894 0.12057637
micoar 528 677 389 3.195909 8054.420 9251.310 0.095596265 0.07777913 0.09573952 0.082168073 0.10559762 0.07026545 0.05952112 0.08733782
cuparu 56 73 49 3.273421 8080.136 9244.732 0.040792615 0.08098799 0.04987732 0.022473972 0.08699604 0.06975892 0.02503962 0.10718172
cha2sc 194 239 190 2.895034 8160.319 9220.697 0.007196491 0.07205628 0.01108164 0.004770191 0.02242886 0.06940662 0.05064604 0.09660343
urerba 2 5 0 3.812822 7954.932 9276.985 Inf 0.24031825 0.13493791 0.022634915 0.35041866 0.06671173 -0.02216408 0.18462937
myrcga 40 50 38 3.231830 8054.830 9266.683 0.015871284 0.06904557 0.02366960 0.009351584 0.04020493 0.06306927 0.02731841 0.10487277
eugega 963 1162 917 3.345195 8044.492 9249.944 0.014631716 0.05615354 0.01599438 0.012605254 0.01824705 0.05595504 0.04743933 0.06617516
$Biggest_losses
N1 N2 S time date1 date2 mortrate little.r fitmort lowermean uppermean fitr lowermeanR uppermeanR
bactc1 241 84 84 3.500939 8002.949 9273.566 0.30105644 -0.30105644 0.29367749 0.26478207 0.3297441 -0.29300499 -0.3248475 -0.22733664
bactba 111 55 55 3.388399 8024.983 9265.000 0.20723565 -0.20723565 0.19885841 0.15353804 0.2639641 -0.19553875 -0.2600445 -0.12258659
pipecu 120 65 62 3.012689 8119.954 9219.893 0.21919198 -0.20350736 0.20422073 0.15152511 0.2847200 -0.18373390 -0.2359292 -0.09849162
bactc2 38 17 17 3.290464 8056.410 9252.538 0.24445571 -0.24445571 0.20608992 0.12881790 0.3019426 -0.17023845 -0.2763264 -0.04498295
hampap 76 49 41 3.217818 8010.668 9258.138 0.19179499 -0.13640085 0.17426848 0.12649105 0.2490157 -0.10871699 -0.1640417 -0.03893491
cecrob 61 38 27 3.123925 8036.937 9260.472 0.26090156 -0.15150417 0.24426914 0.17270408 0.3425667 -0.10431432 -0.1706922 -0.03689557
sennda 201 135 114 3.363347 8028.548 9262.585 0.16861370 -0.11834345 0.16496953 0.14534654 0.2020067 -0.10397926 -0.1475743 -0.05487117
bactma 469 355 341 2.993622 8130.976 9225.702 0.10646643 -0.09302609 0.10539174 0.09120041 0.1183469 -0.08910109 -0.1066423 -0.06859027
conoci 389 279 232 3.555180 7989.365 9270.768 0.14537716 -0.09348825 0.14290808 0.12359480 0.1677055 -0.08625363 -0.1112317 -0.05920075
turpoc 153 113 112 3.229882 8027.949 9258.576 0.09657909 -0.09382700 0.09159398 0.06736020 0.1227418 -0.08600706 -0.1123598 -0.05152248
4. Running the model for several censuses
There is a wrapper function fitSeveralAbundModel that can handle any number of census datasets. It simply fits the abundance model for each pair of censuses, then for the first and last census, saving all into one big list, plus it repeats all the models for larger trees, setting mindbh to 10 times the submitted value. It’s only purpose is to finish a series of models from one plot in a single model run. Since it takes about 30 minutes per census, you might leave it running overnight. The following code saves the results to an R data object in the folder abunddist.
