CTFS Tutorials
Source file: http://ctfs.si.edu/ctfsdev/CTFSRPackageNew/files/tutorials/growthfitAnalysisGrowth vs. DBH
MODELING AGB GROWTH BY OPTIMIZING DBH BINS
Date: April 7, 2011.
The following calculations in R run a model of AGB growth as a function of diameter, or as a function of agb, using a series of discrete size categories and a linear regression within each. The model presented here uses log(dbh) or log(agb) as the predictor (independent variable). The locations of the categories, or bins, of log(dbh) are not defined in advance, but are fitted as part of the model.
The R commands listed show all steps from loading data to running the model and graphing the output. All commands begin with >, and if continued on a second line, +. Comments between the commands describe the input and output of each.
The calculations start with the full data dataframes. These are described at
http://ctfs.arnarb.harvard.edu/Public/CTFSRPackage/help/RoutputFull.pdf, and can be
downloaded from
http://ctfs.arnarb.harvard.edu/yourplot/RAnalyticalTables (of course you have to substitute a plot name
for yourplot).
1. Source necessary functions
The following packages are necessary:
- growth.r
- growthfit.bin.r
- utilities.r
- statistics.r
- distributions.r
Running startCTFS will load these. The argument folder must match the location of the package on your computer.
> startCTFS(folder='/home/condit/CTFSRPackage/')
2. Load plot data
Census data can be loaded using the function CTFSplot in utilities.r (or just attach the files). In the example here, there must be files bci.full5 and bci.full6 inside a folder full inside the listed path (so /home/fullplotdata/full/bci.full5 and same for bci.full6).
3. Calculate growth rates
3.1. Create a table of biomass growth rates using the function extract.growth.
The following creates a table of AGB increment for every individual with dbh≥100 mm. It excludes trees growing faster than 75 mm yr1 (in dbh growth), or shrinking by 4 x the measurement error (which is 1 mm in small trees, 4-6 mm in big trees). Diameter is log-transformed (using logit=’x’). These are appropriate options for the calculations that follow. If you are worried about trimming the outliers, check the functions graph.outliers and graph.outliers.spp in order to see what is excluded. You need to change dbhunit to cm and maxgrow to 7.5 if your dbh is in centimeters, and set mindbh=10.
Of course you should submit census tables from your own plot instead of bci.full5 and bci.full6.
Note that I occasionally use if(!exists). You can ignore these when executing functions. I use it to prevent certain long-running commands from repeating every time I test the calculations. In this case, once agb.growth is created, it won’t be created again.
> cns2=bci.full6
> if(!exists('agb.growth'))
+ agb.growth=extract.growthdata(census1=cns1,census2=cns2,growcol='incgr',
+ growthfunc=growth.biomass.indiv,logit='x',rounddown = FALSE,
+ mindbh = 100,dbhunit = 'mm',err.limit = 4,maxgrow = 75)
19 gustsu 19 5.697093 -0.3739816 0.0071891093
21 virosu 21 5.852202 -0.3032694 0.0098357161
23 quaras 23 5.749393 -0.4840976 -0.0009974601
24 protte 24 6.082219 0.5892221 0.0378020363
25 brosal 25 7.162397 3.1977491 0.0000000000
30 quaras 30 6.146329 0.5224756 0.0465738782
This extracts growth for just one species. You might also make a graph of all growth rates at once in order to explore the data.
> plot(onespdata$dbh,onespdata$growth,pch=16,xlab='log(dbh)',ylab='AGB growth increment')
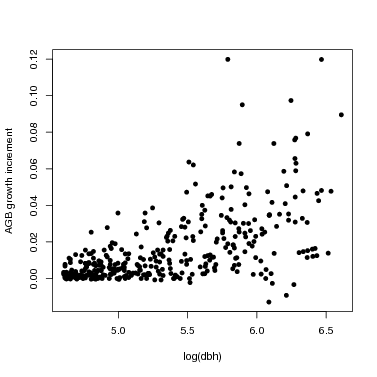
4. The model
Here is how to fit the model for just a single species, then graph the result. The table onespdata produced above has growth data from a single species that is appropriate for the function. For testing, you can start by using method=optim, since it produces the fit faster, but optim does not produce confidence limits, so use method=Gibbs for final results.
Here, dbh is used as the predictor in the model, but this can be changed by setting sizecol=’agb’ instead of dbh.
The option nobin sets how many dbh categories will be used. Here it is set to 2, so just one division point will be estimated. (The location of the division point will be optimized, but the number of division points must be set at the start.)
The option sdmodel defines the model used for related SD to log(size). The default is linear.model.ctr, simply a straight line with intercept defined at the mid-point of the x axis. The number of parameters for the SD model, defined by startsd, must match the number of parameters expected by sdmodel: 2 parameters for a linear model. The second run here (fit2) illustrates a model with 3 parameters, where the SD is constant below a certain value of x, then increases linearly. This allows the SD to increase rapidly at large size while remaining positive at small size.
+ fit=growth.flexbin(growthtable=onespdata,sizecol='dbh',nobin=2,start=NULL,startsd=c(.02,0),
+ sdmodel=linear.model.ctr,method='Gibbs',rep=1500,burn=500,show=100)
> data.frame(names(fit))
1 best
2 bestmean
3 CI
4 bestSD
5 bestSDmean
6 confSD
7 model
8 fullparam
9 sdpar
10 llike
11 burn
12 keep
13 llike.best
14 growth
15 bins
16 optimpar
17 optimSD
18 optimllike
19 max.param
+ fit2=growth.flexbin(growthtable=onespdata,sizecol='dbh',nobin=2,start=NULL,startsd=c(.02,0,.001),
+ sdmodel=constant.linear,method='Gibbs',rep=1500,burn=500,show=100)
The output of the model was saved in the variables fit and fit2, which are lists that include a lot of information. The element fit$model includes the original data (fit$model$x and fit$model$y), plus the fitted value for each (fit$model$pred), and also the fitted SD at every point (fit$model$predSD). The best fit model parameters are fit$best, and confidence limits in fit$CI. The best fit standard deviation parameters are in fit$bestSD. (The model assumes a linear increase in the standard deviation around the line, and the two SD parameters describe the linear increase.)
2.5% 5.182171 0.01109343 0.01427356 0.01025213
97.5% 6.350274 0.01907803 0.13527083 0.01433389
41192 4.605170 0.0024277875 0.002143495 0.002587984
219538 4.605170 0.0033515898 0.002143495 0.002587984
21229 4.615121 0.0067920171 0.002301479 0.002733811
33563 4.615121 0.0077488471 0.002301479 0.002733811
133039 4.615121 0.0002578913 0.002301479 0.002733811
163857 4.615121 0.0007855686 0.002301479 0.002733811
> plot(fit$model$x,fit$model$y,pch=16,xlab='log(dbh)',ylab='AGB growth')
> lines(fit$model$x,fit$model$pred,col='red',lwd=2)
> lines(fit$model$x,fit$model$pred+fit$model$predSD,col='green',lwd=2)
> lines(fit$model$x,fit$model$pred-fit$model$predSD,col='green',lwd=2)
> plot(fit2$model$x,fit2$model$y,pch=16,xlab='log(dbh)',ylab='AGB growth')
> lines(fit2$model$x,fit2$model$pred,col='red',lwd=2)
> lines(fit2$model$x,fit2$model$pred+fit2$model$predSD,col='green',lwd=2)
> lines(fit2$model$x,fit2$model$pred-fit2$model$predSD,col='green',lwd=2)
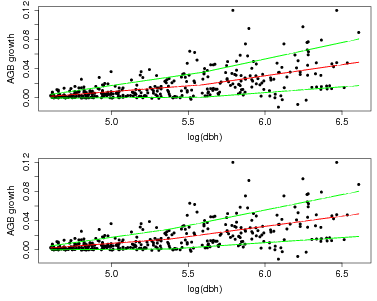
There is a function graph.growthmodel.spp that handles the graphing in a single step. It graphs the points for individual trees, and adds the fitted model in blue (you can change the color). A green line shows the mean growth in 10 equally-spaced divisions (from graphdiv=10). There is also an option to overlay alternative model fits as a way of visualizing statistical confidence in the model, illustrated in the second example below (conf=50): 50 different gray lines are drawn using randomly selected parameters from the Gibbs sampler used to fit the model (all the parameters are returned in fit$fullparam). An advantage of the Gibbs sampler is this way of showing confidence: every set of parameters returned (after the burnin) can be considered equally good fits to the model.
Note that confidence in the model is different than the standard deviation (yellow lines in the previous figure). The latter gives variation among individuals, but the gray lines provide an estimate of confidence in the position of the blue line.
At small dbh (remember, though, only dbh≥ 100 mm are included), the SD is small, meaning little variation among individuals, and the confidence bands are narrow. Both SD and the confidence bands spread at larger size.
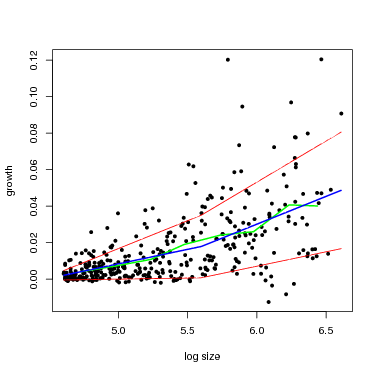
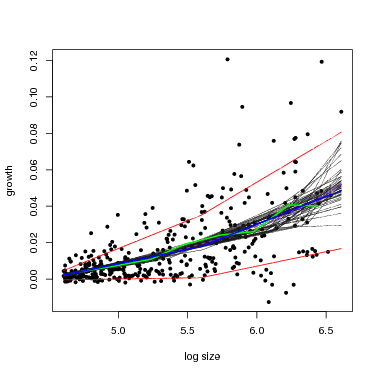
4.3. Interpreting the parameters.
The parameters provided by fit$best describe the model. The first parameter is the break point between the two bins, and the best estimate is 6.04 for in the example here for Tetragastris. The next two parameters are the two slopes, first before the break point, then after the division. In this case, the best fit for them is 0.017 and 0.039, so the slope gets steeper, and both are significantly > 0 (since the confidence intervals do not reach 0). But the two slopes are not significantly different from each other, since their confidence limits overlap. The last parameter is the intercept parameter, which is the estimated growth in the middle of the middle bin.
4.4. Comparing results for different number of bins.
The interesting result is whether using three or four bins improves the model. The function run.growthfit.bin automates the process of running the model 4 times, with 1 through 4 bins. One bin is just a standard linear model; I have not yet tried 5 or more bins.
The following example runs the Gibbs sampler for 1500 repetitions (noreps=1500). In final fits, I recommend using noreps=5000. But for illustration here, the 1500 steps provide adequate parameter values. The variable fitlist creates a list with just one entry, named for the species’ code tet2pa. This is how the data are passed to the graphing function.
+ fit1.4=run.growthfit.bin(growthdata=onespdata,size="agb",startpar=c(.03,.005),
+ startsdpar=c(.04,0),sdmodel=linear.model.ctr,binoption=1:4,
+ noreps=1500,noburn=500,noshow=500)
> fitlist=list(tet2pa=fit1.4)
The funtion overlay.growthbinmodel makes a graph of all 4 models. It expects the fit to be a list, which is why I created fitlist. The first option below shows the graph on the screen; the second exports the graph as a pdf (export=pdf ).
+ modelclr="blue",graphdiv=10,add=FALSE,newgraph=FALSE,export=NULL)
+ modelclr="blue",graphdiv=10,add=FALSE,newgraph=TRUE,
+ export=pdf,outfile="linearbin.overlay.pdf",h=8,w=10)
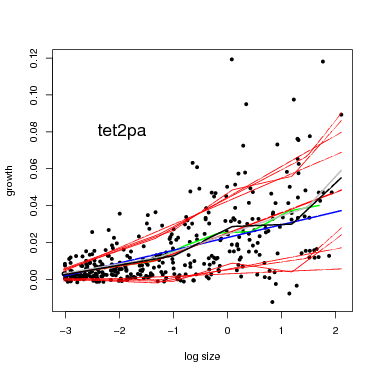
4.5. Fitting the model for all species.
The next step is to run the fit for all species in a plot. Here I limit the selction to species with at least 40 individuals total and 15 individuals ≥300 mm dbh. You may wish to change the limits, but the fitting procedure requires ≥5 individuals per category, so it will fail with species having < 20 individuals above 100 mm dbh.
This will run the Gibbs sampler 5000 steps per species, and will take 10-20 hours to complete. Note that the example here uses agb as the predictor by submitting size=’agb’.
> linearbin.fit.allspp=
+ run.growthbin.manyspp(growthdata=agb.growth,size='agb',spp=allspecies,
+ minabund300=15,minTotal=40,dbhunit='mm',
+ startpar=c(.03,.005),startsdpar=c(.04,0))
> save(linearbin.fit.allspp,file='growth/linearbin.fit.allspp.rdata')
The output, saved in the object linearbin.fit.allspp, is a list, one element per species. Each element has four subelements, one per the models with 1-4 bins. Each of those subelements is a single model fit, with all the elements described above.
The object linearbin.fit.allspp can be passed directly to the function overlay.growthbinmodel to graph results for all the species. In the first example, export=NULL and the graphs are printed to the screen. In the second example, export=pdf and a pdf with all the graphs is created. You can change the file name to whatever you please using outfile.
+ modelclr="blue",graphdiv=10,add=FALSE,newgraph=TRUE,export=NULL)
> overlay.growthbinmodel(fit=linearbin.fit.allspp,bins=1:4,regclr="green",
+ modelclr="blue",graphdiv=10,add=FALSE,newgraph=FALSE,
+ export=pdf,outfile="linearbin.overlay.pdf",h=8,w=10)
4.6. Statistical comparison of the models.
Finally comes a statistical test for assessing the 4 different models (1-4 bins) for each species. The function compare.growthbinmodel calculates the best model using the AIC, or Akaike Information Criteria, and makes a graph of this model.
It also produces a list of 3 different tables:
- The first gives the slope of the growth vs. AGB regression in each bin of the best model. There are up to four slopes per species.
- The second gives the lower 95% confidence limits (or credible intervals in Bayesian terminology) for those same slopes.
- The third has upper credible intervals.
The results from 37 species at BCI are shown below. The columns with NA indicate those slopes were not needed in the best fit, that is, if there is only one slope, then the best model used only one bin, etc. Any negative slope means growth rate declined with size in that size bin. If the upper credible interval is also negative, then the slope is significantly < 0. At BCI, only quaras, Quararibea asterolepis, had a significant decrease in growth rate in the largest size category. Brosimum alicastrum (brosal) and Prioria copaifera (pri2co). had declines, but not significantly so. Poulsenia armata (poular) had a peculiar fit, with a decline in growth in the second to last category, but an increase in the last. Poulsenia was the only species for which all 4 bins were needed, and only 3 other species required 3 bins. More than half the species (20 or 37) were best modeled using a straight line.
Estimated slopes in AGB bins for BCI species, based on the model with the highest AIC.
> model.compare$slopes
alchco 0.0032465338 NA NA NA
alsebl 0.0021806654 0.007128261 0.10890872 NA
apeime 0.0046786758 0.035849494 NA NA
aspicr 0.0283336937 NA NA NA
beilpe 0.0033653161 0.013751175 NA NA
brosal 0.0024068701 0.047129124 -0.17796505 NA
casear -0.0253325121 0.008690913 NA NA
cecrin 0.0050180792 NA NA NA
cordal 0.0117999289 NA NA NA
cordbi 0.0039835569 NA NA NA
dendar 0.0025569194 0.116643536 NA NA
drypst 0.0036403207 0.018269066 NA NA
guapst 0.0103665438 NA NA NA
guargu 0.0019973357 NA NA NA
guatdu -0.0010994482 0.027943355 NA NA
gustsu 0.0015635930 NA NA NA
heisco 0.0011742999 NA NA NA
hirttr 0.0017723195 NA NA NA
huracr 0.0161435081 1.183355785 NA NA
jac1co 0.0145098911 NA NA NA
loncla 0.0073481019 NA NA NA
luehse 0.0079207901 NA NA NA
ocotwh 0.0147476415 NA NA NA
poular 0.0008762754 0.049263099 -0.08280719 0.1826693
poutre 0.0150712909 NA NA NA
pri2co 0.0028850771 0.033175754 -0.02125288 NA
protte 0.0017907969 NA NA NA
quaras 0.0067436907 -0.033460006 NA NA
simaam 0.0059474071 0.027965169 NA NA
sponra 0.0047933671 0.040976050 NA NA
tab1ro 0.0069016656 0.166647884 NA NA
tab2ar 0.0077546177 NA NA NA
tet2pa 0.0069909722 NA NA NA
tri2tu 0.0039848927 0.012648412 NA NA
virose 0.0023402584 NA NA NA
virosu -0.0060751517 0.017998711 NA NA
zantbe 0.0241310891 NA NA NA
Upper confidence limits for slopes in AGB bins. Any of these < 0 indicate a slope significantly less than zero. There are also lower confidence limits (not shown), and any of those > 0 indicate a significantly positive slope.
alchco 0.005792440 NA NA NA
alsebl 0.003896948 0.008517255 0.162687781 NA
apeime 0.005269493 0.116905396 NA NA
aspicr 0.035465859 NA NA NA
beilpe 0.004758150 0.016956525 NA NA
brosal 0.005181650 0.066313528 0.032898032 NA
casear -0.000402689 0.012374577 NA NA
cecrin 0.006276610 NA NA NA
cordal 0.016520575 NA NA NA
cordbi 0.005075125 NA NA NA
dendar 0.004150830 0.168619765 NA NA
drypst 0.004727390 0.027774678 NA NA
guapst 0.013970905 NA NA NA
guargu 0.002744795 NA NA NA
guatdu 0.002862032 0.068154978 NA NA
gustsu 0.001816538 NA NA NA
heisco 0.001714928 NA NA NA
hirttr 0.002388317 NA NA NA
huracr 0.022292374 1.485571198 NA NA
jac1co 0.016943170 NA NA NA
loncla 0.009518829 NA NA NA
luehse 0.014597148 NA NA NA
ocotwh 0.020653555 NA NA NA
poular 0.001844633 0.065679098 -0.008801082 0.2599029
poutre 0.017404537 NA NA NA
pri2co 0.005328017 0.040479278 0.010913174 NA
protte 0.002503612 NA NA NA
quaras 0.007552048 -0.006168187 NA NA
simaam 0.008965739 0.039581944 NA NA
sponra 0.010183281 0.085681413 NA NA
tab1ro 0.012633114 0.461891000 NA NA
tab2ar 0.009206953 NA NA NA
tet2pa 0.007870435 NA NA NA
tri2tu 0.004513265 0.017689708 NA NA
virose 0.002991947 NA NA NA
virosu 0.012829836 0.038371756 NA NA
zantbe 0.032282730 NA NA NA
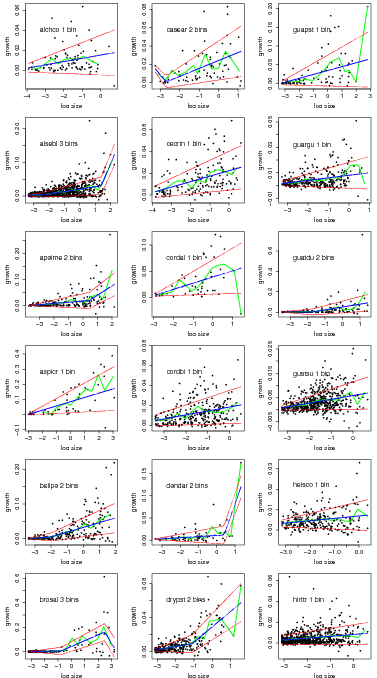
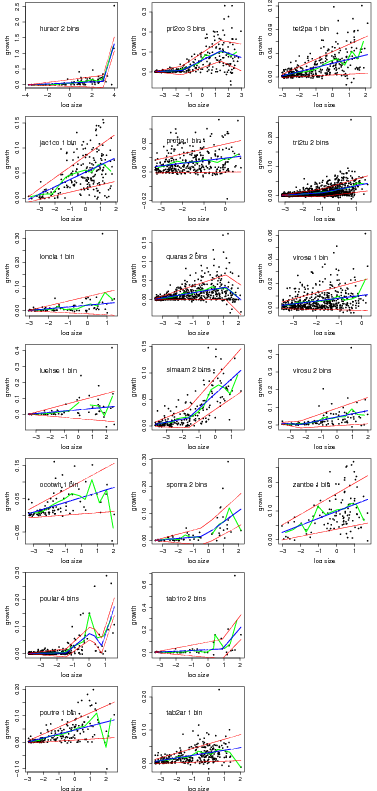
Graphs of the best model based on AIC for all species.
> temp=compare.growthbinmodel(fit=linearbin.fit.allspp[1:18],export=NULL,makegraph=TRUE,
+ newgraph=FALSE,conf=0)
> temp=compare.growthbinmodel(fit=linearbin.fit.allspp[19:37],export=NULL,makegraph=TRUE,
+ newgraph=FALSE,conf=0)
Now use a run with an updated trimming of extreme growth, permitting more negative growth (see trimGrowth), and the alternative model of SD where the slope can turn upward at large size.
+ trimmed3=extract.growthdata(census1=cns1,census2=cns2,growcol='incgr',growthfunc=growth.biomass.indiv,
+ logit='x',rounddown = FALSE,pomcut=.05,mindbh = 100,
+ dbhunit = 'mm',err.limit = 12,maxgrow = 40)
> if(!exists('linearbin.fit.trim3'))
+ linearbin.fit.trim3=
+ run.growthbin.manyspp(growthdata=trimmed3,size='agb',spp=allspecies,minabund300=15,
+ minTotal=40,dbhunit='mm',startpar=c(.03,.005),startsdpar=c(.04,0,.001),
+ badsdfunc=bad.binsdpar,sdmodel=constant.linear)
> model.compare=compare.growthbinmodel(fit=linearbin.fit.trim3,export=NULL,makegraph=FALSE)
> model.compare$slopes
alchco 2.313291e-03 NA NA NA
alsebl 8.801125e-04 0.006729967 0.1165375 NA
apeime 5.377643e-03 NA NA NA
aspicr 7.659387e-03 0.056497966 NA NA
beilpe 3.434838e-03 0.014251442 NA NA
brosal -9.937160e-06 0.045653429 NA NA
casear 6.236175e-03 NA NA NA
cecrin 4.344572e-03 NA NA NA
chr2ar 4.342516e-04 NA NA NA
cordal 1.119337e-02 NA NA NA
cordbi 3.850902e-03 NA NA NA
dendar 3.061239e-03 0.103986771 NA NA
drypst 4.046738e-03 NA NA NA
guapst 8.774790e-03 NA NA NA
guargu 1.933300e-03 0.016178454 NA NA
guatdu -3.233411e-04 0.012647192 NA NA
gustsu 1.484349e-03 NA NA NA
heisco 1.037609e-03 NA NA NA
hirttr 1.787888e-03 NA NA NA
huracr 7.576259e-04 0.076699153 0.1477084 NA
ingama 1.218236e-02 NA NA NA
jac1co 1.558190e-02 NA NA NA
loncla 4.174202e-03 NA NA NA
luehse 6.472729e-03 NA NA NA
ocotwh 1.638501e-02 -1.507224977 NA NA
poular 1.232570e-03 0.036611405 NA NA
poutre 5.097094e-03 0.017228969 NA NA
pri2co 3.300787e-03 0.025710413 NA NA
protte 1.633665e-03 NA NA NA
quaras 6.211712e-03 NA NA NA
simaam 6.996646e-03 0.029710478 NA NA
sponra 3.689212e-03 0.038736659 NA NA
tab1ro 7.552564e-04 NA NA NA
tab2ar 7.830165e-03 NA NA NA
tachve 4.926326e-03 0.128908115 NA NA
tet2pa 6.873703e-03 NA NA NA
tri2tu 2.661230e-03 0.008013637 NA NA
virose 2.286633e-03 NA NA NA
virosu 7.480564e-04 0.019794913 NA NA
zantbe 1.879699e-02 NA NA NA
alchco 0.0047805797 NA NA NA
alsebl 0.0026159073 0.007851163 0.1842141 NA
apeime 0.0071833650 NA NA NA
aspicr 0.0191826934 0.080957890 NA NA
beilpe 0.0057810590 0.017617172 NA NA
brosal 0.0006205874 0.073233290 NA NA
casear 0.0086761343 NA NA NA
cecrin 0.0063719731 NA NA NA
chr2ar 0.0024420043 NA NA NA
cordal 0.0153462792 NA NA NA
cordbi 0.0052195064 NA NA NA
dendar 0.0051950156 0.176018295 NA NA
drypst 0.0052035559 NA NA NA
guapst 0.0130986123 NA NA NA
guargu 0.0028043138 0.069607345 NA NA
guatdu 0.0023154959 0.018383915 NA NA
gustsu 0.0018811924 NA NA NA
heisco 0.0016746418 NA NA NA
hirttr 0.0023245174 NA NA NA
huracr 0.0028343234 0.107355627 0.6054979 NA
ingama 0.0223377262 NA NA NA
jac1co 0.0174474237 NA NA NA
loncla 0.0073316976 NA NA NA
luehse 0.0107542164 NA NA NA
ocotwh 0.0222512241 -0.322498697 NA NA
poular 0.0018242545 0.051693888 NA NA
poutre 0.0140629751 0.025875535 NA NA
pri2co 0.0051542918 0.031276790 NA NA
protte 0.0024564011 NA NA NA
quaras 0.0070888172 NA NA NA
simaam 0.0096095343 0.040682545 NA NA
sponra 0.0076408366 0.091596492 NA NA
tab1ro 0.0038115804 NA NA NA
tab2ar 0.0092720217 NA NA NA
tachve 0.0116175257 0.250464577 NA NA
tet2pa 0.0082352449 NA NA NA
tri2tu 0.0039823663 0.009757620 NA NA
virose 0.0029423020 NA NA NA
virosu 0.0063641490 0.063036954 NA NA
zantbe 0.0256678498 NA NA NA
Graphs of the best model based on AIC for all species.
> temp=compare.growthbinmodel(fit=linearbin.fit.trim3[1:21],export=NULL,makegraph=TRUE,
+ newgraph=FALSE,conf=0)
> temp=compare.growthbinmodel(fit=linearbin.fit.trim3[22:40],export=NULL,makegraph=TRUE,
+ newgraph=FALSE,conf=0)
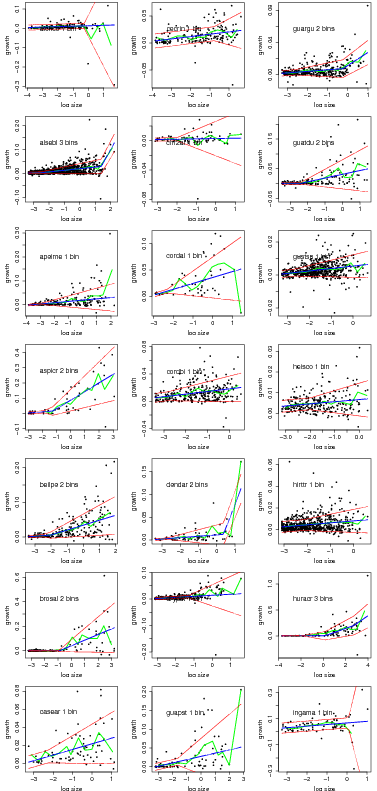
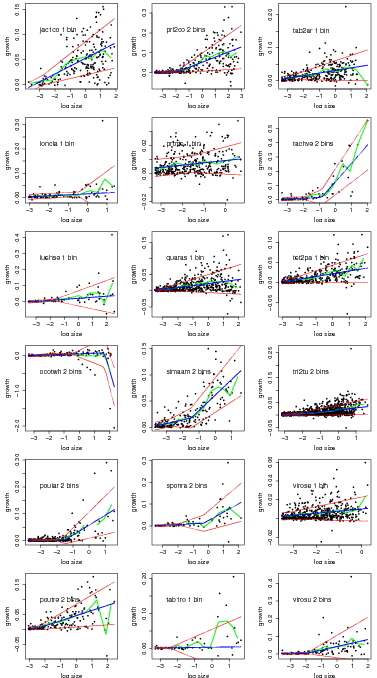
You can also export all the graphs at once into a single pdf to a file named by outfile.
+ newgraph=FALSE,outfile="linearbin.orig.pdf")
> temp=compare.growthbinmodel(fit=linearbin.fit.trim3,export=pdf,makegraph=TRUE,
+ newgraph=FALSE,outfile="linearbin.trim3.pdf")
5. File creation
Anyone intereted in seeing how Sweave works in R to create this document can contact me.
> system("pdflatex growthFitBin");
> system("htlatex growthFitBin");
> system("rm ~/data/growth/growthfitAnalysis/*")
> system("mv ~/data/growthFitBin* ~/data/growth/growthfitAnalysis/");
> system("mv ~/data/growth/growthfitAnalysis/growthFitBin.html ~/data/growth/growthfitAnalysis/index.html");
> system("rsync -ave ssh ~/data/growth/growthfitAnalysis rcondit@ctfs.arnarb.harvard.edu:/var/www/html/Public/CTFSRPackage/tutorials/ --whole-file");
> system("cp -r ~/data/growth/growthfitAnalysis /var/www/Public/CTFS_RPackage/files/tutorials/")
